Tutorial 1: Protein - DNA interaction
- Note:
- This tutorial uses the current Biskit snapshot from the subversion repository (after-2.1)
- You find the python code and PDB file in biskit/docs/tutorial1
- For a quicker overview, you can simply read / execute meganuclease.py which also contains some comments
- I've slightly run out of steam for the last part of the tutorial -- send me a mail if anything is unclear
1. Open and inspect the structure
Open an interactive python shell (type python or open the python file in emacs and start a new interpreter) and execute the following:
from Biskit import *
m0 = PDBModel('1R7M.pdb')
The first line imports all published Biskit classes, including PDBModel.
Warning: you should never use 'import *' in your own python programs. For interactive work it is acceptable though.
The second line creates a new instance of PDBModel and loads the example PDB file into it. Now, let's see what we can do with this PDBModel thing! First we want to have a look at the sequence:
m0.sequence() 'NIKKNQVMNLGPNSKLLKEYKSQLIELNIEQFEAGIGLILGDAYIRSRDEGKTYCMQFEWKNKAYMDHVCLLYDQWVLSPPHKKER VNHLGNLVITWGAQTFKHQAFNKLANLFIVNNKKTIPNNLVENYLTPMSLAYWFMDDGGKWDYNKNSTNKSIVLNTQSFTFEEVEYL VKGLRNKFQLNCYVKINKNKPIIYIDSMSYLIFYNLIKPYLIPQMMYKLPNIKKNQVMNLGPNSKLLKEYKSQLIELNIEQFEAGIG LILGDAYIRSRDEGKTYCMQFEWKNKAYMDHVCLLYDQWVLSPPHKKERVNHLGNLVITWGAQTFKHQAFNKLANLFIVNNKKTIPN NLVENYLTPMSLAYWFMDDGGKWDYNKNSTNKSIVLNTQSFTFEEVEYLVKGLRNKFQLNCYVKINKNKPIIYIDSMSYLIFYNLIK PYLIPQMMYKLPacgctagggataacagggtaatacgtattaccctgttatccctagcgtacgctagggataacagggtaatacgta ttaccctgttatccctagcgt++++++~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~'
Apparently, we are looking at a pretty large structure here! First comes a protein sequence (upper case letters) followed by a stretch of DNA residues (lower case). It's difficult to make out much more. What, for instance, are those + and ~ characters? Do we have one or two DNA strands? PDBModel.report() [New after release 2.1.0] gives you a more useful overview:
m0.report()
[PDBModel 1R7M 5883 atoms, 749 residues, 8 chains]
* chain 0: NIKKNQVMNLGPNSKLLKEYKSQLIELNIEQFEAGIGLILGDAYIRSRDEGKTYCMQF..
* chain 1: NIKKNQVMNLGPNSKLLKEYKSQLIELNIEQFEAGIGLILGDAYIRSRDEGKTYCMQF..
* chain 2: acgctagggataacagggtaatac
* chain 3: gtattaccctgttatccctagcgt
* chain 4: acgctagggataacagggtaatac
* chain 5: gtattaccctgttatccctagcgt
* chain 6: ++++++
* chain 7: ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~..
source: LocalPath[ {/home/raik|$HOME}/data/papers/2007_10_paris/1R7M.pdb ]
15 atom profiles: ['name', 'residue_number', 'type', 'alternate', 'name_o..
0 residue profiles: []
1 info records: ['date']
Now things become a bit clearer. We are looking at two, apparently identical, protein chains and 4 DNA strands. The data from the PDB have ended up in 15 different "atom profiles". Let's check those out:
m0.atoms
ProfileCollection: 15 profiles of length 5883
name
{'isarray': False, 'default': None, 'version': '18/11/2007 ProfileCollection
$Revision: 559 $; PDBModel $Revision: 560 $', 'changed': 0}
['N', 'CA', 'C', 'O', 'CB', 'CG', 'OD1', 'ND2'...
residue_number
{'isarray': True, 'default': 0, 'version': '18/11/2007 ProfileCollection
$Revision: 559 $; PDBModel $Revision: 560 $', 'changed': 0}
[ 3 3 3 ..., 199 200 201]
temperature_factor
{'isarray': True, 'default': 0, 'version': '18/11/2007 ProfileCollection
$Revision: 559 $; PDBModel $Revision: 560 $', 'changed': 0}
[ 61.95000076 61.61999893 61.38000107 ..., ...
Mhm... that's a lot of information (and I've cut away the remaining 12 profiles). I'll quickly walk you through. So you are basically looking at something like a dictionary of lists (on steroids). The keys describe the columns of the PDB file:
m0.atoms.keys() ['name', 'residue_number', 'type', 'alternate', 'name_original', 'chain_id', 'occupancy', 'element', 'segment_id', 'charge', 'residue_name', 'after_ter', 'serial_number', 'insertion_code', 'temperature_factor']
Each entry is a list (array actually) with one value for each atom in the PDB. Whatever you do with your PDBModel (see below), each value in each profile is always guaranteed to stay aligned (or perish together) with the atom it belongs to. Each profile also carries a bunch of meta-data (you can add your own).
OK, so how do you get to the values? There are different ways. Let's first have a look at the B-Factor profile of this structure:
t = m0.atoms['temperature_factor'] import Biskit.gnuplot as G G.plot( t )
This should pop up a plot like this:

:
The gnuplot module has not actually been written by us. We have included a copy in Biskit to spare you (or rather us) a separate installation and, more importantly, because later versions of this python module are more powerful but less easy to use. We use the biggles module for any serious plotting but there is nothing quite as simple as gnuplot if you just want to have a quick look at a bunch of numbers.
2. Inspecting atoms & residues
The above is a 'profile centric' view on the data -- so we are looking how a certain value behaves along all atoms. m0.atoms also offers a 'atom-centric' view:
m0.atoms atom = m0.atoms[-1]
The above lines extract a Pseudo-dictionary holding every single values of all atom profiles for the last atom of our PDB file (the first atom would be atoms[0]):
atom
CrossView{'name': 'O', 'residue_number': 201, 'insertion_code': '', 'alternate': '',
'name_original': ' O ', 'chain_id': '', 'occupancy': 1.0, 'element': 'O', 'segment_id': '',
'charge': '', 'residue_name': 'HOH', 'after_ter': 0, 'serial_number': 5889, 'type': 'HETATM',
'temperature_factor': 52.3899993896}
Now you can guess the meaning of those "~" symbols in m0.sequence() above. "~" is used for any residue that Biskit recognizes as solvent. The "+" stands for positive and "-" for negative ions -- if recognized, otherwise, you will see a "?" for unknown residues.
The 'CrossView' behaves like a dictionary. There is an important difference though: The atom CrossView stays connected to its underlying profiles. Any change to any of the profiles' last value will be mirrored in atom and, vice-versa, you can change the model's profiles by changing the values of a CrossView:
## this evaluates to False m0.atoms['temperature_factor'][-1] == 0 False ## change last profile position (via the CrossView) atom['temperature_factor'] = 0 m0['temperature_factor'][-1] == 0 True
Did you notice that there is no ".atoms" in the last statement above? PDBModel defines several shortcuts so that you rarely have to type "m0.atoms" but can directly index the model:
## The following statements are all True
m0[-1] == m.atoms[-1]
m0['temperature_factor'] == m0.atoms['temperature_factor']
## You can also iterate over all atom CrossViews:
for atom in m0:
print atom['name'], ': ', atom['occupancy'], ' / ', atom['temperature_factor']
The idea is that PDBModel can, at first approximation, be treated as a list of atoms. So the number of atoms in a PDBModel is considered its length:
len( m0 ) 5883
Why do we then have this .atoms thing at all? Remember, there are also residue profiles for residue-centered data:
## explore residue profiles m0.residues ProfileCollection: 0 profiles of length 0
Well, as you see, freshly loaded from a PDB file, PDBModel actually doesn't have any residue profiles. But we can easily create some:
## create profile with single-character Amino acid code per residue m0['aa'] = m0.sequence() ## or convert the atom profile of residue names into a residue profile m0.residues['rname'] = m0.atom2resProfile( 'residue_name' )
:
Note for advanced hackers:
The last statement is equivalent to:
import numpy as N m0['rname'] = N.take( m0['residue_name'], m0.resIndex() )That means, atom2resProfile creates a residue profile from the value of each residue's first atom. But you can also use the residue average or provide your own 'averaging' function. See m0.atom2resProfile.__doc__!
But wait a minute! Why would a simple "m0['aa'] = m0.sequence()" work? Shouldn't it be m0 .residues ['aa'] = m0.sequence()? You are right! But as mentioned, we have defined short cuts. PDBModel looks at the list of values from which you want to create a profile. If it has the same length as there are atoms, PDBModel will create an atom profile. If instead the length corresponds to the number of residues, as in this case, we end up with a new residue profile. If the length doesn't match either, you will receive an exception.
3. Extract the biological unit
The m0.report() above indicated that we may have some duplicates in the PDB
m0.report( plot=1 ) ## calls m0.plot()
...will additionally give you a quick and dirty pseudo-graphical overview of the structure you are looking at:

Clearly, we are not going to win a molecular graphics competition with this one (We will produce some more competitive Pymol images below). But it's enough to see that we are dealing with two copies of a protein-DNA complex.
Note for hackers: m0.plot is simply plotting all x against all y coordinates. You can do that yourself in one step:
G.plot( zip( m0.xyz[:,0], m0.xyz[:,1] ) )You see the advantages of keeping all the data in straight numpy arrays?
Judging from the text output of report() (see above), our biological unit would comprise the first protein chain and, presumably, the first two DNA chains. Let's extract those into a new PDBModel instance m:
m = m0.takeChains( [0, 2, 3 ] )
The result looks like this:
m.plot()

Looks like it worked!
4. Analyze protein-DNA contacts
Let's first calculate and visualize a distance matrix of all against all atoms:
import Biskit.mathUtils as MU pw = MU.pairwiseDistances( m.xyz, m.xyz ) ## nicer graphics with Biskit.Matrixplot p = MatrixPlot( pw, step=25 ) p.show()
MatrixPlot (thanks to Wolfgang Rieping!) uses the biggles plotting library. With biggles we are approaching publication quality:

Blue spots picture low, red large distances. Do you see the cross formed by the two anti-parallel DNA strands? Anyway, most people are more used to binary "contact plots". Since we already have a matrix in numpy format, these are easy to produce:
cont = N.less( pw, 10 ) G.scatter( zip( *N.nonzero( cont ) ) )
numpy.less gives us a two-dimensional mask with '1' in every position where the pw value is below 10 and 0 (or False) in all other positions. We fell back to gnuplot for plotting the contacts (MatrixPlot becomes excessively slow for more than 100 x 100 data points). scatter() is the gnuplot method for scatter plots (surprise!) and expects a list of (x,y) tuples. What we have is a mask though. So we have extracted the position of each '1' with numpy.nonzero and unpacked (that's why the *) the result into the zip method. Well, maybe, for sanity, you want to split that up into more than one line...
We don't really care about all those contacts within the protein or within the DNA. So let us only look at intermolecular contacts that form the protein DNA interface! First we separate the DNA from the protein:
mask_dna = [ name in ['A','C','T','G'] for name in m['residue_name'] ] dna = m.compress( mask_dna ) ## write a new PDB file so that you can inspect the DNA from outside dna.writePdb( 'dna_only.pdb' ) prot = m.compress( m.maskProtein() )
We have first created a mask (an array of 0 and 1) with 1 for each DNA atom ('name in ...' evaluates to either True or False). We have then given this mask to PDBModel.compress which returns a new PDBModel instance only containing the atoms marked with 1 (or True). Then we did the same for the protein part. Just that we didn't have to create the mask by hand -- PDBModel already comes with a handy function for that (maskProtein, there also is maskCA, maskBB, maskHeavy, etc.).
Now is a good time to introduce a more specialized Biskit module. The Biskit.Dock package bundles classes for handling complexes:
from Biskit.Dock import Complex com = Complex( prot, dna )
We now have created a Biskit.Dock.Complex from two PDBModels. What methods does Complex offer us?:
dir( com ) ['atomContacts', 'compareAtoms', 'compress', 'conservationScore', 'contPairScore', 'contactResDistribution', 'contactResPairs', 'contactTypeDistribution', 'contacts', 'contactsDiff', 'contactsOverlap', 'contactsShared', 'equalAtoms', 'extractLigandMatrix', 'foldXEnergy', 'fractionNativeContacts', 'fractionNativeSurface', 'get', 'getInfo', 'has_key', 'info', 'initVersion', 'interfaceArea', 'keys', 'lig', 'ligMatrix', 'lig_model', 'lig_transformed', 'ligandMatrix', 'loadResContacts', 'model', 'prosa2003Energy', 'pw_dist', 'rec', 'rec_model', 'resContacts', 'resPairCounts', 'rmsInterface', 'rmsLig', 'rtTuple2matrix', 'setLigMatrix', 'slim', 'take', 'values', 'version', 'writeComplex', 'writeLigand', 'writeReceptor']
Alright, there is lot's of stuff. You better have a look at the API documentation for this class. Several methods deal with contact matrices:
cont = com.atomContacts( cutoff=10 ) N.shape( cont ) (1854, 984) print cont [[0 0 0 ..., 0 0 0] [0 0 0 ..., 0 0 0] [0 0 0 ..., 0 0 0] ..., [0 0 0 ..., 0 0 0] [0 0 0 ..., 0 0 0] [0 0 0 ..., 0 0 0]]
Numpy spares us the view of all the 1854 by 984 data points (1854 protein versus 984 DNA atoms). Obviously most positions are 0 (no contact). The plot looks interesting though:

5. Visualization
Biskit contains a simple wrapper for the pymol program. The following code snippet colors each atom by the number of its intermolecular contacts:
# how many contacs has each dna and protein atom? contacts_lig = N.sum( cont, axis=0 ) contacts_rec = N.sum( cont, axis=1 ) pm = Pymoler() pm.addPdb( com.rec(), 'rec' ) pm.addPdb( com.lig(), 'lig' ) pm.colorAtoms( 'rec', contacts_rec ) pm.colorAtoms( 'lig', contacts_lig ) pm.add( 'show surface') pm.run()
If you have Pymol installed, you should now be looking at this:

|

|
| Pymol window | protein with DNA switched off |
6. Add and visualize conservation data
It may be interesting to compare the contact density with the conservation of a residue. Conservation profiles can be extracted with the Hmmer program from Pfam alignments. This is automated in Biskit.Hmmer. PDBDope is a convenience class that maps results from many external programs into PDBModel profiles:
prot = com.rec_model
d = PDBDope( prot )
d.addConservation()
## This will take a while, or load the pre-calculated result:
## prot = T.Load('rec.model')
prot.residues
pm = Pymoler()
pm.addPdb( prot, 'rec' )
pm.addPdb( com.lig(force=1), 'lig')
pm.colorRes( 'rec', prot['cons_ent'] )
pm.add( 'show surface')
pm.run()
Will yield this:
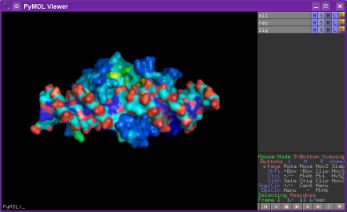
|

|
| Pymol window | protein with DNA switched off |
7. Correlate conservation and contact values
So there is some increased conservation in the interface but is there really a correlation? There is a little problem though: The conservation profile is a residue profile. Our contact profile has one value for each atom rather than residue. So we first need to create a residue profile from that and then we can plot the two profiles against each other:
prot['rescontacts'] = prot.atom2resProfile( 'contacts', f=N.sum ) ## scatter plot of raw profiles G.scatter( zip( prot['rescontacts'], prot['cons_ent'] ) ) ## line plot of normalized profiles _cont = prot['rescontacts'] / 1. / N.max( prot['rescontacts'] ) _cons = prot['cons_ent'] / 1. / N.max( prot['cons_ent'] ) G.plot( _cont, _cons )
Note:
'f=N.sum' means that the value for a residue will be calculated from the atom values using the function numpy.sum. You can write your own function and pass it as an argument. The default is numpy.average.

|
scatter plot |

|
line plot |
It doesn't look as if there is any direct correlation between the conservation of a residue and the number of its contacts. However, the protein analyzed here has an engineered interface. The natural homing endonuclease may show a better signal. We leave this question for you to explore...
So this was a brief crash-course to some of Biskit's core functionality. Comments and questions are welcome!
